One of the most visited post of all times in this blog is the OOmmf post with a quick introduction into OOmmf and the comparison between simulation and a real magnetic material images. That’s the reason for this post. I think people will like it.

Here we are going to create a simulation, store the data and create nice animations with it. And on top of that, we are going to learn how to export the simulation data into Matlab and Python.
Let’s start selecting the structure we want to simulate and the parameters.
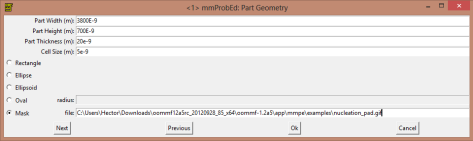
The mask we are going to use is this one (click on it to download, and remember to put it into your ….\oommf12a5rc_20120928_85_x64\oommf-1.2a5\app\mmpe\examples folder).
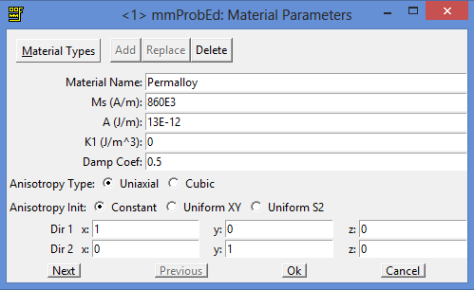
 And the material is going to be Permalloy.
And the material is going to be Permalloy.
This structure is interesting because it’s a typical structure to study Domain Wall (DW) nucleation and propagation. When we ramp the magnetic field from left to right we are going to see how a DW forms on the big pad and moves towards the needle-like end on the right.
So. Let’s go with the geometrical parameters for this structure. We are going to select them in a way that the nanowire connecting the pad with the sharp end is going to be around 150 nanometers wide. So, according to the image size, the mask is going to be 700 nanometers height and 3800 nanometers wide. The cell size is going to be 5 nanometers (for Permalloy the exchange length is 5 nanometers, so the cell must be that size or even smaller, and many published papers use 5 nanometer in the simulations).
For the material we are going to select Permalloy as it comes in the program with standard parameters.
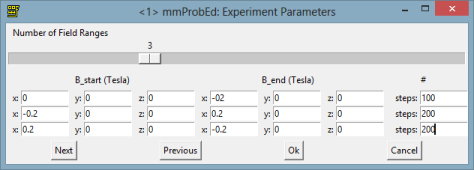
And now let’s specify the external magnetic field during the simulation. We are going to create 3 steps where the field is changing in this way:
from 0 to -200mT in x axes
from -200 to 200 in x axes
from 200 to -200 in x axes
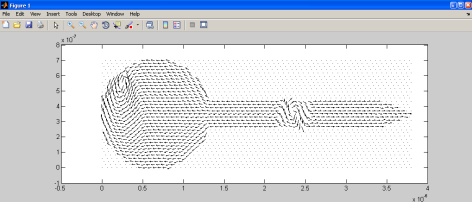
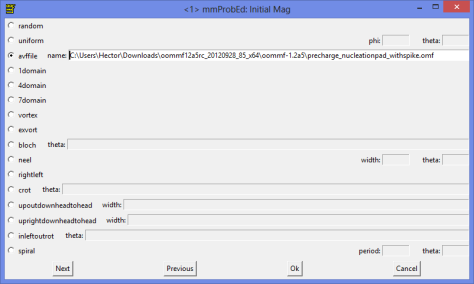
The next thing is now to select the initial magnetization. You can use a random magnetization. In our case we are going to use a previous saved file which magnetization divergence looks like this:
Just load the data file into the initial magnetization path.
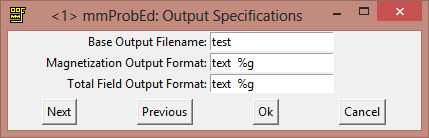
Later we will explain how to save data, because this is a very useful trick to resume simulations or start from a particular state. Now the final step to prepare the problem is to select the output and change it from binary 4 to text %g. In this way the magnetization in each point will be stored as a text number. In this way is going to be quite easy to load the data into Matlab to do more analysis.
We are ready now to do the simulations, the next thing is to open a display to show the magnetization on the screen and 2 archives to save data. Remember to keep all of them on the screen, including the problem editor. Sometimes if you close the problem editor the values are reset.
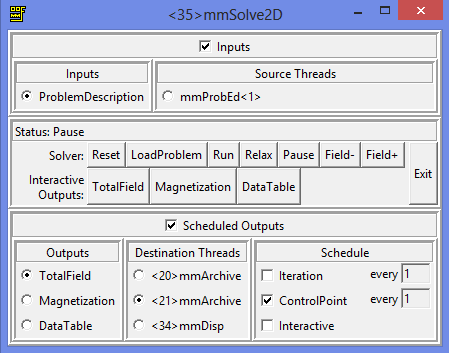
Ok, so click Solve2D, load the problem and select to visualize the magnetization on the display every 100 iterations. For the first archive select magnetization and store also every 100 iterations. For the second archive store total field every 100 iterations. Ideally we only want to save every control point, that is, when a stable magnetization is reached for every magnetic field step. But the Domain Wall propagation along the wire is a dynamic process, and we will miss it if we store on control points (I know, I did it before). And why 100? Why not every one iteration? Because I did this simulation before and it is going to take about 406934 iterations, so even saving every 100 means more than 4000 data files.
This is going to generate screen outputs every 100 iteration steps, and save the data on every control point (each time equilibrium is reached for a set field).
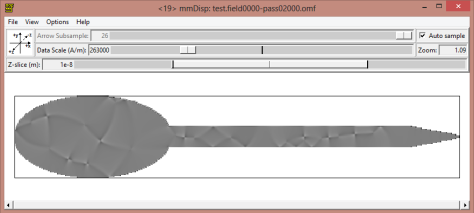
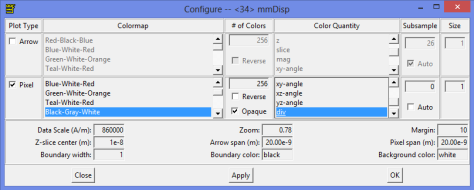
If you want to display the data while running the simulation as we do here, this is the configuration for the display. A little trick is to change the data scale on the display and reduce it. That will increase the contrast on the image.
.
.
.
It took me a few days finishing the simulation (not running during nigths and not at full speed (that is something I need to investigate why)), but finally you will have 1004 files generated. Half of them will look like test.field0xxx-passxxxxxx.ohf that ones are for the total field on each point of the simulation, and half of them will look like test.field0xxx-passxxxxxx.omf that ones are for the magnetization at each point.
ok. Can we actually transform these files into something we can “see” or analyse in some way? Yes, we can.
We have:
About 500 files with magnetization (*.omf).
About 500 files with total field (*.ohf).
Our options
Turn Data into Images and compose an animation.
Import data into another program and do an animation there.
Turn Data into Images and compose an animation.
Let’s try the easy route. Turn data into Images and compose an animation. To do that, on the display window File>Show Console and we start to figth with the OOmmf console. I say this because some commands will work, others not… and everything will sound weird to people like me, used to write commands in other way. Anyway. After some try and error I found that this command will turn all the .omf files in the main OOmmf folder into jpg images.
tclsh oommf.tcl avf2ppm *.omf -config magnetic.txt -opatsub .jpg -filter "tclsh oommf.tcl any2ppm -format jpeg"
You just type it in and it will turn all the magnetization files into jpeg images. But before doing it, remember to create a options file named (in this case) magnetic.txt It will be a text file with options for the conversion. This is how it looks like in my case.
array set plot_config {
colormaps { Red-Black-Blue Blue-White-Red Teal-White-Red \
Black-Gray-White White-Green-Black Red-Green-Blue-Red }
arrow,status 0
arrow,colormap Black-Gray-White
arrow,colorcount 0
arrow,quantity z
arrow,autosample 1
arrow,subsample 10
arrow,size 1
arrow,antialias 1
pixel,status 1
pixel,colormap Black-Gray-White
pixel,colorcount 60
pixel,quantity div
pixel,autosample 0
pixel,subsample 1
pixel,size 1
misc,background #FFFFFF
misc,drawboundary 1
misc,margin 10
misc,width 1280
misc,height 960
misc,crop 1
misc,zoom 0
misc,rotation 0
misc,datascale 13200
}
The whole explanation for these commands is in the Nist webpage here.
You can do the same with the total field data, but is quite boring. Just change omf for ohf in the comand and select arrows in the configuration file.
Once the magnetization images are created, we can remove the first 100 (remember that those correspond to the first ramping up of the field from zero to high x negative values) and the remaining 400 images can be used to create a loop in a video, because the last one and the first one will have the same magnetization and also the same external field.
To create an animated GIF. This time we are going to use GIMP.
 Just go to the webpage, download and install, it’s free and I will tell you how to use it for this.
Just go to the webpage, download and install, it’s free and I will tell you how to use it for this.
Open the program. And File>Open as layers and select all the images you have just created (first order them by date). They will appear on the layer menu and the first one will be show on the display.
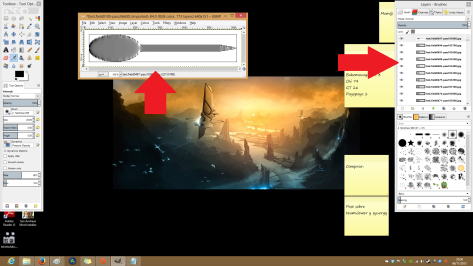
And now is as simple as File>Export and select GIF on the type of file… remember to add .gif after the file name. And select export as animation.
And this is how it looks like (with low quality).

NICE!!!! It looks amazing. Notice the two kinds of Domain Walls, one when the magnetic field is saturating to the left and another one when it is saturating to the right. Also, on the pad, where there is some roughness on the surface, you can also see the Domain Wall moving like jumping, the Barkhausen effect (Domain Wall encountering defects on its way and getting pinned to them)
Now let’s try the other option.
Import data into another program and do an animation there.
To do that, I have selected Matlab and I wrote a code to import the data into it. Can be found here at the Matlab File Exchange.
Basically, what the code does is read the data files line by line, extract the data like size of the simulation, number of points used, magnetic field… And then, because we select the output to be text %g it just reads the coordinate of each vector on the vector field. To have an idea this is how a data file looks like (You can open *.omf files with Notepad and this will show).
# OOMMF: rectangular mesh v1.0 # Segment count: 1 # Begin: Segment # Begin: Header # Title: C:/Users/Hector/Downloads/oommf12a5rc_20120928_85_x64/oommf-1.2a5/test.field0000-pass03369.omf # Desc: Field Index: 0 # Desc: Applied field (T): 0 0 0 # Desc: Iteration: 3369 # Desc: Time (s): 1.8747579070869034e-9 # Desc: |m x h|: 0.092583929421348854 # Desc: User Comment: # meshtype: rectangular # meshunit: m # xbase: 2.5e-009 # ybase: 2.5e-009 # zbase: 1e-008 # xstepsize: 5e-009 # ystepsize: 5e-009 # zstepsize: 2e-008 # xnodes: 400 # ynodes: 100 # znodes: 1 # xmin: 0 # ymin: 0 # zmin: 0 # xmax: 2e-006 # ymax: 5e-007 # zmax: 2e-008 # valueunit: A/m # valuemultiplier: 800000 # ValueRangeMinMag: 1e-8 # ValueRangeMaxMag: 1.0 # End: Header # Begin: Data Text -0 0 0 0 0 -0 -0 0 -0 -0.263872 -0.417373 0.000337065 -0.238721 -0.433818 0.000330522 -0.211667 -0.449099 0.000320794 -0.182926 -0.462911 0.000308174 -0.152747 -0.474998 0.000293057 -0.121395 -0.485153 0.000275874 # End: Data Text # End: Segment
I cut down part of the data to make it fit. Notice the vectors because they have 3 coordinates, but notice they are not in the place they occupy in the simulation. To arrange them in the correct order you need to extract the data of number of x y and z nodes.
If you want to download and try my code, do it. Once you have the data on Matlab the limit is your imagination. This is how some of it will look alike.
And this is all for now. Hope you like it. Next time more.

Hey there, you have done some interesting things with OOMMF. I was just wondering if you know any ways in which to examine OOMMF 3d output in Matlab, as I would like to look at net magnetization for models I’ve created, but can’t seem to open up files.
Hi Matt.
Somebody reported me the same problem a few days ago. I fixed and submitted a new script to file exchange. It will be approved by the moderators shortly. I will let you know when it is. (12 hours from now maybe).
Thanks.
Héctor.
It is now available the new version.
Hey again,
I tried the solution but my models are unfortunately too big to open in Matlab….
Do you know how to tell mmArchive (perhaps through the mif file) to write its output in text format as opposed to binary?
I would like to open these output files (created after each stage) in Excel, in order to calculate total magnetization and plot hysteresis graphs, but unfortunately at the moment it isn’t working nicely….
Hi Matt.
If it is a 2D problem, I told how to get the output in text in this post. You need to go to the problem editor, in the “Output Specifications” and change “binary 4” for “text %g”.
For 3D problems I haven’t work with them yet, but I made a search. Here you can see a MIF example (scroll down the webpage).
http://math.nist.gov/oommf/doc/userguide12/userguide/MIF_2.1.html#sec:mif2format
Find these 2 lines in the MIF example.
scalar_output_format “%#.8g”
vector_field_output_format {binary 4}
I guess changing “binary 4” for “text %g” will work.
About using Excel instead of Matlab… I know that Matlab has a limit to the size of it’s arrays. Probably Excel has this limit too and probably they will be smaller. Let me know how it works please.
(I can tell you now that there is a script for Python to convert binary files into Matlab arrays, but is not as clean as using only Matlab).
You can also use OOMMF comand line tools avf2ovf or avf2odt to convert existing OVF files from binary to text formats. For example,
tclsh oommf.tcl avf2ovf -format text infile.ovf outfile.ovf
Produces data lines that look like
-0.89075910999999997 -0.45418781000000003 0.016176810000000000
-0.70800065999999995 -0.70620048000000002 0.0040030246999999998
0.0056123388999999996 -0.99991297999999995 -0.011937468999999999
Here you still need to figure out the point locations from the header info, as discussed in the article. If you prefer you can use the ‘-grid irreg’ option to get embedded position info; in that case the data lines look like
0.0000000000000000 0.0000000000000000 0.0000000000000000 -0.89075910999999997 -0.45418781000000003 0.016176810000000000
1.0000000000000000 0.0000000000000000 0.0000000000000000 -0.70800065999999995 -0.70620048000000002 0.0040030246999999998
2.0000000000000000 0.0000000000000000 0.0000000000000000 0.0056123388999999996 -0.99991297999999995 -0.011937468999999999
The ‘-grid irreg’ option is nice for importing into other programs, but OOMMF itself mostly doesn’t want -grid irreg files, and there is no provision in OOMMF for converting from -grid irreg back to -grid rect.
The output format for avf2ovf text is %.17g, which retains full 8-byte (double) floating point precision. Alternatively, avf2odt makes text data tables from OVF files and allows you to specify the format for numeric values. avf2odt will do various types of averaging (for example, you can get cross-sectional averages), but the following example uses the -average point option to get a one-to-one rendering of the input data:
tclsh oommf.tcl avf2odt -average point -numfmt %g infile.ovf
which writes by default to the file named “infile.odt” with lines that look like
0 0 0 -890759 -454188 16176.8
1 0 0 -708001 -706200 4003.02
2 0 0 5612.34 -999913 -11937.5
Note the difference in magnitude between the avf2ovf output and the avf2odt output. The OVF format includes in the header a “valuemultiplier” setting, which in this case has the value 1000000. You need to multiply the field values in OVF files by the valuemultiplier setting to get the true value.
Links to avf2ovf and avf2odt documenation:
http://math.nist.gov/oommf/doc/userguide12a5/userguide/Vector_Field_File_Format_Co.html
http://math.nist.gov/oommf/doc/userguide12a5/userguide/Making_Data_Tables_from_Vec.html
Re: fighting with the OOMMF console
The OOMMF console is not documented very well. Although, as discussed in the article, you can use the console to run OOMMF commands, you can also run OOMMF commands from your usual command shell. The example
tclsh oommf.tcl avf2ppm *.omf -config magnetic.txt -opatsub .jpg
-filter “tclsh oommf.tcl any2ppm -format jpeg”
will work fine (except see below) from any of the usual unix shells (e.g., bash). The Windows command prompt does not do wildcard expansion, so on Windows you need to use “-ipat *.omf” instead of just “*.omf”. (This is explained in the avf2ppm documentation.)
The OOMMF console is actually just a souped-up Tcl/Tk prompt—it is essentially the Windows version of the Wish console. If you know Tcl then the console is a convenient place to debug OOMMF and try out snippets of Tcl code. If you aren’t very familiar with Tcl you are probably better off running OOMMF command apps from your usual shell.
I also wanted to note that the ‘any2ppm -format jpeg’ command may not work on some systems. any2ppm will convert to any format that the underlying Tk installation supports. IIRC, PPM, BMP, and GIF support are guaranteed, but beyond that you need additionalTcl/Tk packages installed, such as the Img package. You can see if Img is installed on your system by bringing up the OOMMF console or a Wish prompt, and entering the command “package require Img”. If it is available the result will be the Img package version number. Some Tcl/Tk distributions automatically include Img (I think ActiveTcl does), otherwise you can install Img separately. On Linux/Debian-based systems the package seems to be called “libtk-img”; on rpm-based systems I’ve seen both tkimg and tk-Img.
Hi, out of curiosity, is there anyway you can modify size of region within Imageatlas function? For example, on imageatlas, there is a section of
colormap {
color-1 region_name
color-2 region_name
…
color-n region_name
}
where we can define each colors as certain region, I was wondering if those region size can be modified to have different thickness for example.
Thank you very much for any response
Hi,
Can any one tell me, how to draw a 3D image for array of cylinders. I use 3D – OOMMF 1.2a5. I have also converted .omf from binary4 to text format. I am not able to see the 3D view of cylindrical array using the text file.
Hi Rajan
I have never solved a 3D problem, only 2D, so I’m probably not the best one to answer. Where are you trying to represent the results? Are you using my script to export vector files into Matlab?
I want to find the size of the magnetic domain from the min driver magnetisation image obtained from mm display of oxsii by using MATLAB. Can you help me for that code? I wrote my own, but it is showing error.
Hi. I will try. What is the error about? please, tell me more.
Has anyone tried creating an array of close packed spheres? (naturally a 3d problem)
I would like to create a solid which would resemble an FCC lattice, with planes of spheres, however at the moment I can’t think of how to get them close packed, rather the plane above line zero starts at the upper surface of the sphere below which is a rather rough approximation.
Thanks in advance if anyone can suggest any solutions to this.
My problem is that I never used the 3D tool, so I cannot really give you a proper answer. But try to simulate 2 circles one close to the other, maybe the interaction is very weak for your material parameters and the spheres can be simulated as independent from each other.
Can you please tell me the total command for PBC_Exchange_2D in OOMMF oxii?? It is showing some error on that line.
Hi. I never use that module. I suppose the best way to proceed is contacting the authors. http://math.nist.gov/oommf/contrib/oxsext/
Inside the module’s files there is an example, maybe that can help you.
http://sourceforge.net/projects/oommf-2dpbc/files/?source=navbar
Please answer … its a kind of urgent. Any link or suggestion will also work.